Loading BLAST Annotations¶
Creating An Analysis¶
To load a blast analysis, navigate to Content > Tripal Content. At the top of the page, click Add Tripal Content and select Analysis from the list of content types. Some sites may have custom analysis types for each type of analysis performed. For our dataset, we need to make two analyses: one for TrEMBL and one for Swiss-Prot.
(note: the above step is optional, but recommended).
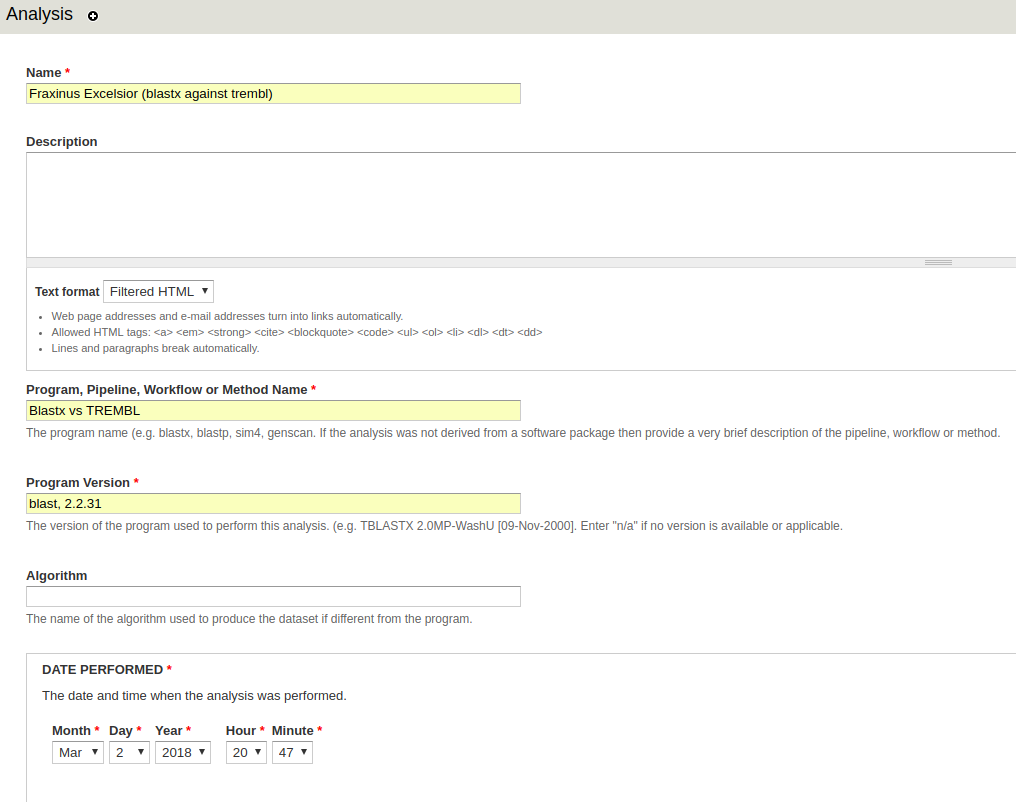
Enter data into the following fields:
- Analysis Name - The name should be <organism common name> (<blast version> against <database>). For example American Chestnut (blastx against sprot).
- Program, Pipeline Name or Method Name - Note that as of the time that this is being written, an analysis will not be saved if this field matches the Program, Pipeline Name or Method Name of an existing analysis. For this reason, it’s recommend that you use Blastx vs Swiss-Prot for sprot and Blastx vs TREMBL for TREMBL.
- Program, Pipeline or Method version - Something along the lines of blast, 2.2.31.
- Date Performed - This should be the date the blast was run. If the blast process of scripts took several days, use the first day the job was created. If no date can be ascertained, then use 01, 01, 1900.
- Data Source Name - This will be the name of the unigene. There is not really a standard for a source that is a whole genome (like Chinese chestnut).
- Data Source Version - This is the version of the unigene or assembly. This field is optional and may be left blank.
Other fields may be left at their default values or empty. Click save.
Loading BLAST Results¶
The BLAST loader is handled by the tripal_analysis_blast module. The BLAST loader can only load data from blast results in XML format.
Locate the BLAST loader from the menu through Tripal > Data Loaders > Chado BLAST XML Results Loader.
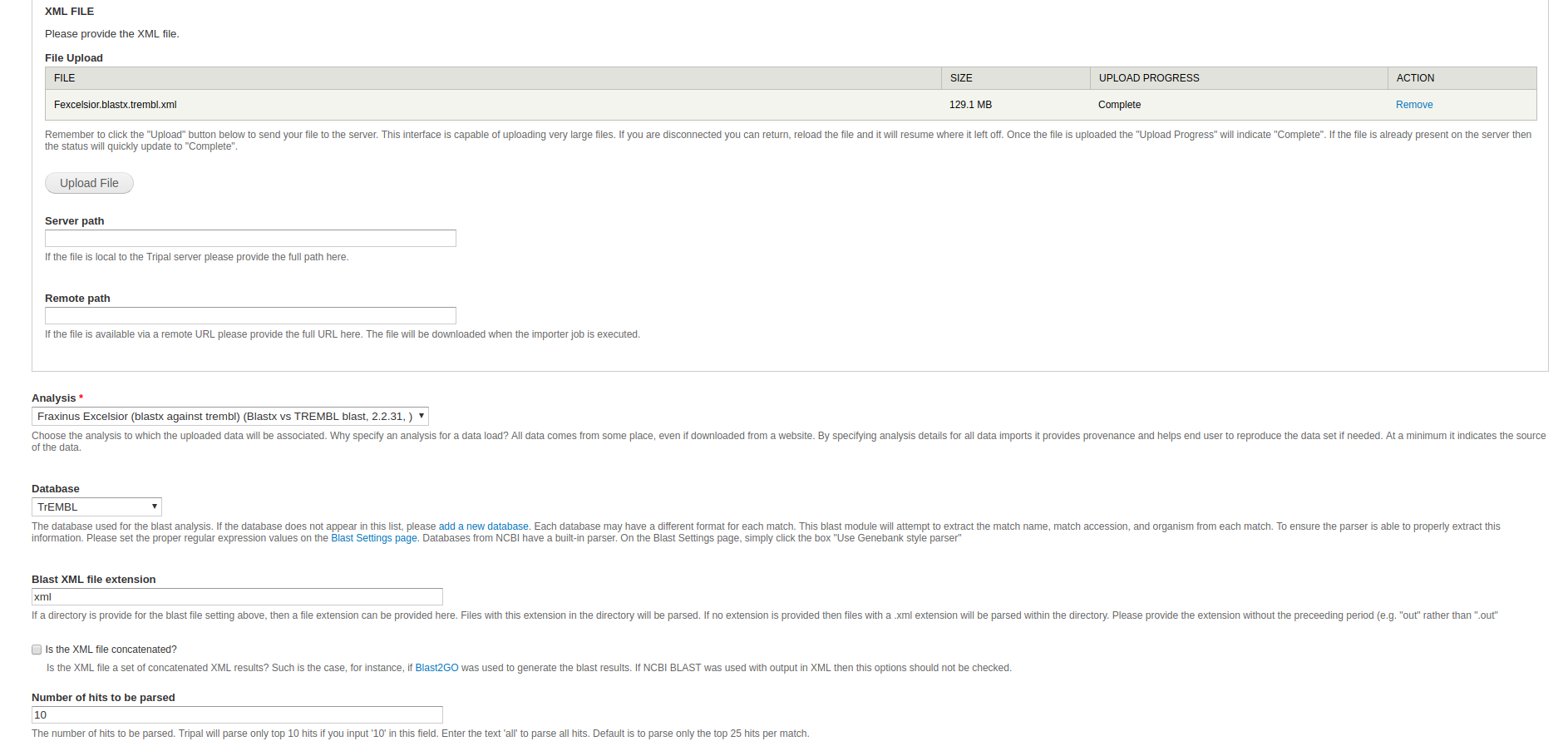
- XML File - Select and upload a blast xml file or provide a path to the blast xml file. If you are using a path, do not provide the file extension. If you input a directory without the tailing slash, all xml files in the directory will be loaded.
- Analysis - Select the newly created blast analysis.
- Database - You will need to create database entries for Swiss-prot and TrEMBL. Select the database that corresponds to the XML file you’re loading (i.e. Swiss-prot for sprot and trembl for trembl).
- Blast XML File Extension - If you provided a path to the xml file instead of uploading a file directly, this would be the time to specify the file extension. This would typically be set to xml.
- Number of hits to be parsed - Set this value to 10.
Other fields may be left at their default values or empty. Click Import File at the bottom of the page. You will need to run the job provided.
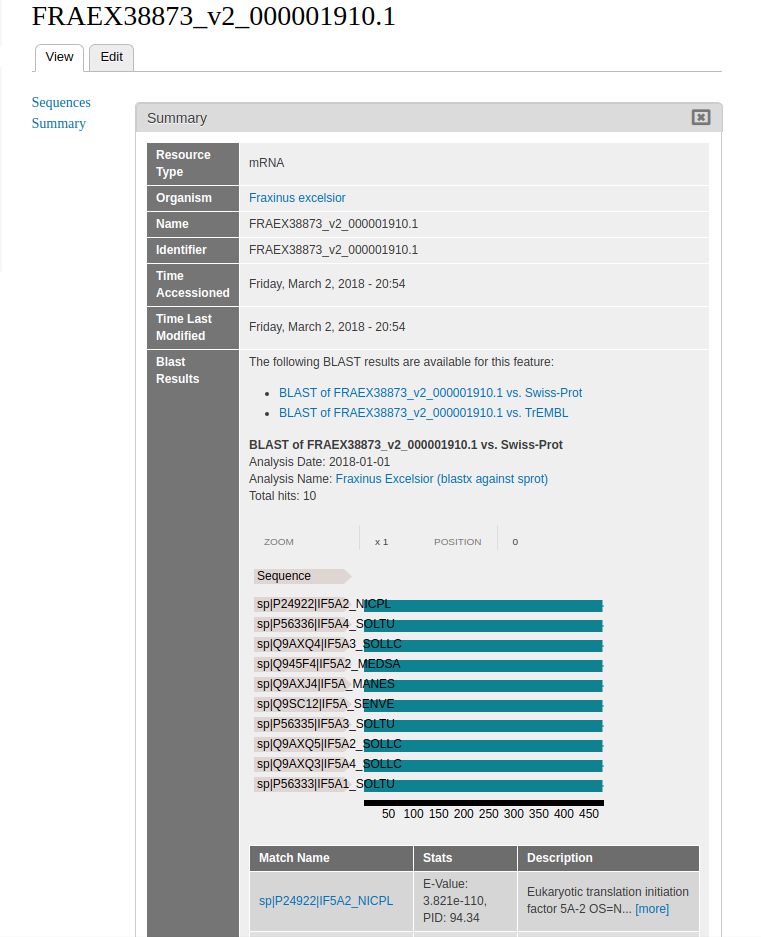
Viewing BLAST Results¶
Most fields are not enabled by default: this includes the BLAST results field. In order for the BLAST results to show up on mRNA entities, we must enable the field.
From the menu, navigate to Structure > Tripal Content Types. If the field format__blast_display is not listed, you should press the “Check for new fields” button in the upper left, and the field should be automatically added (but disabled by default). In the new window, select manage display in the table next to the content type mRNA.
At the bottom of this window is a field of disabled content types, under which Blast Results should be located. Drag Blast Results out of the disabled field.
Blast results should now be viewable in any mRNA content.